Research in life sciences is only possible today with access to online databases. Extracting information
useful for medical researchers and practitioners is possible now with the methods of parallel data mining,
simultaneously applied to high throughput data, molecular databases, and medical publications.
Mining Mass Spectrometric/Proteomics Data
High throughput mass spectrometry analysis produces large amounts of noisy data that have to be filtered and preprocessed with computational tools before subjected to detailed analysis and interpretation. Our strategy uses principles borrowed from cognitive psychology for identifying patterns in mass spectra. Namely, the human
mind is able to capture holistic features in complex sensory inputs, and we trust that similar principles can be applied to abstract data structures. The bioinformatics support of proteomics research is a central theme in our projects. We develop new tools capable of filtering and processing large data streams characteristic of high
throughput analysis workflows.
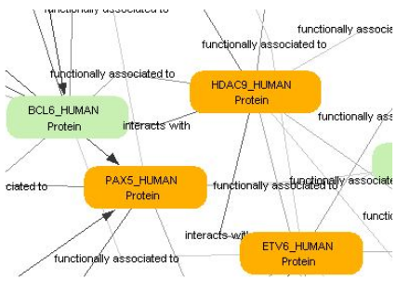
Medical hypothesis generation
Hypothesis generation refers to generating surprising, non-trivial suppositions, and explanations based on information extracted from textual resources. From a data mining perspective, text-based hypothesis generation is a case of link discovery, i.e. a hypothesis can be considered as an undiscovered relation between pre-existing knowledge items. Early success stories include the discovery of therapies for Raynaud’s disease and migraine. In the genomics era, hypotheses are often formulated as relations involving molecular entities, such as genes, proteins, drugs, metabolites, etc., so the use of textual resources needs to be combined with molecular databases, and often, with new experimental data generated by the user. A typical example of application is finding undiscovered links and synergisms between approved pharmaceuticals, as drug combinations, can reach the applications phase much faster than novel drugs. A promising area is the study of synergisms that may exist between generic and targeted therapeutic agents or the design of cocktail therapies for complex diseases.
The emphasis of current cancer therapy is shifting from traditional chemotherapy to targeted drugs. Such therapies rest on two fundamental motives: i) the use of targeted pharmacons that act on one or a few molecular targets specific to tumor cells, and ii) identification of biomarkers suitable for the prediction of drug response. High throughput technologies provide massive amounts of data that can be processed from many viewpoints, the average research groups however lack the necessary and sometimes very extensive, bioinformatics repertoire. Our aim is to develop on-line facilities that are able to integrate high throughput data with complex algorithmic procedures that allow identification of biomarkers or statistical targets. An additional goal is to create prediction systems that can help point of care diagnostics applications.
Project Participants:
- Balázs Ligeti, PhD student
- Prof. Sándor Pongor, PI
Collaborators:
- Dr. Balázs Győrffy
Research Laboratory of Pediatrics and Nephrology, Hungarian Academy of Sciences
Semmelweiss University, Budapest, Hugary - Beáta Reiz
Szeged Biological Centre, Szeged, Hungary - Dr. Ingrid Petrič
Centre for Systems and Information Technologies
University of Nova Gorica, Slovenia - Dr. Mike Myers
International Centre for Genetic Engineering and Biotechnology, Trieste, Italy - Dr. Attila Kertész-Farkas
International Centre for Genetic Engineering and Biotechnology, Trieste, Italy
References
2542344
{2542344:ITV6C94W},{2542344:XYTW8HTD},{2542344:ZMJY9KY8},{2542344:SN743IDY},{2542344:3LSJDGCV},{2542344:FXGP8HQJ},{2542344:LA252I2N},{2542344:2IVYXPJI},{2542344:4VTZS4QB},{2542344:H4YVYQ6N},{2542344:YCIIGX2J},{2542344:VRCKVGQ9},{2542344:TDKLYCSX}
items
1
apa
0
default
asc
188
https://pongor2.itk.ppke.hu/wp-content/plugins/zotpress/
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3A%22zotpress-be75cb866ffea9fee25a04a1173c417e%22%2C%22meta%22%3A%7B%22request_last%22%3A0%2C%22request_next%22%3A0%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%222IVYXPJI%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Cserh%5Cu00e1ti%20et%20al.%22%2C%22parsedDate%22%3A%222011-05%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ECserh%26%23xE1%3Bti%2C%20M.%2C%20Tur%26%23xF3%3Bczy%2C%20Z.%2C%20Zombori%2C%20Z.%2C%20Cserz%26%23xF6%3B%2C%20M.%2C%20Dudits%2C%20D.%2C%20Pongor%2C%20S.%2C%20%26amp%3B%20Gy%26%23xF6%3Brgyey%2C%20J.%20%282011%29.%20Prediction%20of%20new%20abiotic%20stress%20genes%20in%20Arabidopsis%20thaliana%20and%20Oryza%20sativa%20according%20to%20enumeration-based%20statistical%20analysis.%20%3Ci%3EMolecular%20Genetics%20and%20Genomics%3A%20MGG%3C%5C%2Fi%3E%2C%20%3Ci%3E285%3C%5C%2Fi%3E%285%29%2C%20375%26%23x2013%3B391.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00438-011-0605-4%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2Fs00438-011-0605-4%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Prediction%20of%20new%20abiotic%20stress%20genes%20in%20Arabidopsis%20thaliana%20and%20Oryza%20sativa%20according%20to%20enumeration-based%20statistical%20analysis%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22M%5Cu00e1ty%5Cu00e1s%22%2C%22lastName%22%3A%22Cserh%5Cu00e1ti%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Zolt%5Cu00e1n%22%2C%22lastName%22%3A%22Tur%5Cu00f3czy%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Zolt%5Cu00e1n%22%2C%22lastName%22%3A%22Zombori%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mikl%5Cu00f3s%22%2C%22lastName%22%3A%22Cserz%5Cu00f6%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22D%5Cu00e9nes%22%2C%22lastName%22%3A%22Dudits%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22J%5Cu00e1nos%22%2C%22lastName%22%3A%22Gy%5Cu00f6rgyey%22%7D%5D%2C%22abstractNote%22%3A%22Plants%20undergo%20an%20extensive%20change%20in%20gene%20regulation%20during%20abiotic%20stress.%20It%20is%20of%20great%20agricultural%20importance%20to%20know%20which%20genes%20are%20affected%20during%20stress%20response.%20The%20genome%20sequence%20of%20a%20number%20of%20plant%20species%20has%20been%20determined%2C%20among%20them%20Arabidopsis%20and%20Oryza%20sativa%2C%20whose%20genome%20has%20been%20annotated%20most%20completely%20as%20of%20yet%2C%20and%20are%20well-known%20organisms%20widely%20used%20as%20experimental%20systems.%20This%20paper%20applies%20a%20statistical%20algorithm%20for%20predicting%20new%20stress-induced%20motifs%20and%20genes%20by%20analyzing%20promoter%20sets%20co-regulated%20by%20abiotic%20stress%20in%20the%20previously%20mentioned%20two%20species.%20After%20identifying%20characteristic%20putative%20regulatory%20motif%20sequence%20pairs%20%28dyads%29%20in%20the%20promoters%20of%20125%20stress-regulated%20Arabidopsis%20genes%20and%2087%20O.%20sativa%20genes%2C%20these%20dyads%20were%20used%20to%20screen%20the%20entire%20Arabidopsis%20and%20O.%20sativa%20promoteromes%20to%20find%20related%20stress-induced%20genes%20whose%20promoters%20contained%20a%20large%20number%20of%20these%20dyads%20found%20by%20our%20algorithm.%20We%20were%20able%20to%20predict%20a%20number%20of%20putative%20dyads%2C%20characteristic%20of%20a%20large%20number%20of%20stress-regulated%20genes%2C%20some%20of%20them%20newly%20discovered%20by%20our%20algorithm%20and%20serve%20as%20putative%20transcription%20factor%20binding%20sites.%20Our%20new%20motif%20prediction%20algorithm%20comes%20complete%20with%20a%20stand-alone%20program.%20This%20algorithm%20may%20be%20used%20in%20motif%20discovery%20in%20the%20future%20in%20other%20species.%20The%20more%20than%201%2C200%20Arabidopsis%20and%201%2C700%20Orzya%20sativa%20genes%20found%20by%20our%20algorithm%20are%20good%20candidates%20for%20further%20experimental%20studies%20in%20abiotic%20stress.%22%2C%22date%22%3A%22May%202011%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1007%5C%2Fs00438-011-0605-4%22%2C%22ISSN%22%3A%221617-4623%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-28T14%3A05%3A12Z%22%7D%7D%2C%7B%22key%22%3A%22FXGP8HQJ%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Reiz%20et%20al.%22%2C%22parsedDate%22%3A%222014-06-01%22%2C%22numChildren%22%3A1%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EReiz%2C%20B.%2C%20P.%20Myers%2C%20M.%2C%20Pongor%2C%20S.%2C%20%26amp%3B%20Kert%26%23xE9%3Bsz-Farkas%2C%20A.%20%282014%29.%20Precursor%20Mass%20Dependent%20Filtering%20of%20Mass%20Spectra%20for%20Proteomics%20Analysis.%20%3Ci%3EProtein%20and%20Peptide%20Letters%3C%5C%2Fi%3E%2C%20%3Ci%3E21%3C%5C%2Fi%3E%288%29%2C%20858%26%23x2013%3B863.%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Precursor%20Mass%20Dependent%20Filtering%20of%20Mass%20Spectra%20for%20Proteomics%20Analysis%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Be%5Cu00e1ta%22%2C%22lastName%22%3A%22Reiz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michael%22%2C%22lastName%22%3A%22P.%20Myers%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Attila%22%2C%22lastName%22%3A%22Kert%5Cu00e9sz-Farkas%22%7D%5D%2C%22abstractNote%22%3A%22Identification%20and%20elimination%20of%20noise%20peaks%20in%20mass%20spectra%20from%20large%20proteomics%20data%20streams%20simultaneously%20improves%20the%20accuracy%20of%20peptide%20identification%20and%20significantly%20decreases%20the%20size%20of%20the%20data.%20There%20are%20a%20number%20of%20peak%20filtering%20strategies%20that%20can%20achieve%20this%20goal.%5CnHere%20we%20present%20a%20simple%20algorithm%20wherein%20the%20number%20of%20highest%20intensity%20peaks%20retained%20for%20further%20analysis%20is%20proportional%20to%20the%20mass%20of%20the%20precursor%20ion.%20We%20show%20that%20this%20technique%20provides%20an%20improvement%20over%20other%20intensity%20based%20strategies%2C%20especially%20for%20low%20mass%20precursors.%22%2C%22date%22%3A%222014-06-01%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22ISSN%22%3A%22%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-20T22%3A01%3A49Z%22%7D%7D%2C%7B%22key%22%3A%223LSJDGCV%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kert%5Cu00e9sz-Farkas%20et%20al.%22%2C%22parsedDate%22%3A%222012%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKert%26%23xE9%3Bsz-Farkas%2C%20A.%2C%20Reiz%2C%20B.%2C%20Myers%2C%20M.%20P.%2C%20%26amp%3B%20Pongor%2C%20S.%20%282012%29.%20Database%20Searching%20In%20Mass%20Spectrometry%20Based%20Proteomics.%20%3Ci%3ECurrent%20Bioinformatics%3C%5C%2Fi%3E%2C%20%3Ci%3E7%3C%5C%2Fi%3E%282%29%2C%20221%26%23x2013%3B230.%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Database%20Searching%20In%20Mass%20Spectrometry%20Based%20Proteomics%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Attila%22%2C%22lastName%22%3A%22Kert%5Cu00e9sz-Farkas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Be%5Cu00e1ta%22%2C%22lastName%22%3A%22Reiz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michael%20P.%22%2C%22lastName%22%3A%22Myers%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%5D%2C%22abstractNote%22%3A%22%22%2C%22date%22%3A%222012%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22ISSN%22%3A%22%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-10T12%3A21%3A17Z%22%7D%7D%2C%7B%22key%22%3A%22LA252I2N%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Reiz%20et%20al.%22%2C%22parsedDate%22%3A%222012-06-01%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EReiz%2C%20B.%2C%20Kert%26%23xE9%3Bsz-Farkas%2C%20A.%2C%20Pongor%2C%20S.%2C%20%26amp%3B%20Myers%2C%20M.%20P.%20%282012%29.%20Data%20Preprocessing%20and%20Filtering%20in%20Mass%20Spectrometry%20Based%20Proteomics.%20%3Ci%3ECurrent%20Bioinformatics%3C%5C%2Fi%3E%2C%20%3Ci%3E7%3C%5C%2Fi%3E%282%29%2C%20212%26%23x2013%3B220.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.2174%5C%2F157489312800604363%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.2174%5C%2F157489312800604363%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Data%20Preprocessing%20and%20Filtering%20in%20Mass%20Spectrometry%20Based%20Proteomics%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Be%5Cu00e1ta%22%2C%22lastName%22%3A%22Reiz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Attila%22%2C%22lastName%22%3A%22Kert%5Cu00e9sz-Farkas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michael%20P.%22%2C%22lastName%22%3A%22Myers%22%7D%5D%2C%22abstractNote%22%3A%22Mass%20spectrometry%20based%20proteomics%20analysis%20can%20produce%20many%20thousands%20of%20spectra%20in%20a%20single%20experiment%2C%20and%20much%20of%20this%20data%2C%20frequently%20greater%20than%2050%25%2C%20cannot%20be%20properly%20evaluated%20computationally.%20Therefore%20a%20number%20of%20strategies%20have%20been%20developed%20to%20aid%20the%20processing%5Cnof%20mass%20spectra%20and%20typically%20focus%20on%20the%20identification%20and%20elimination%20of%20noise%2C%20which%20can%20provide%20an%20immediate%20improvement%20in%20the%20analysis%20of%20large%20data%20streams.%20This%20is%20mostly%20carried%20out%20with%20proprietary%20software.%20Here%20we%20review%20the%20current%20main%20principles%20underlying%20the%20preprocessing%5Cnof%20mass%20spectrometry%20data%20give%20an%20overview%20of%20the%20publicly%20available%20tools.%22%2C%22date%22%3A%222012-06-01%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%2210.2174%5C%2F157489312800604363%22%2C%22ISSN%22%3A%22%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.benthamdirect.org%5C%2Fpages%5C%2Fb_viewarticle.php%3FarticleID%3D3177023%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-10T12%3A21%3A06Z%22%7D%7D%2C%7B%22key%22%3A%22XYTW8HTD%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Petri%5Cu010d%20et%20al.%22%2C%22parsedDate%22%3A%222014-06-01%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EPetri%26%23x10D%3B%2C%20I.%2C%20Ligeti%2C%20B.%2C%20Gy%26%23x151%3Brffy%2C%20B.%2C%20%26amp%3B%20Pongor%2C%20S.%20%282014%29.%20Biomedical%20Hypothesis%20Generation%20by%20Text%20Mining%20and%20Gene%20Prioritization.%20%3Ci%3EProtein%20and%20Peptide%20Letters%3C%5C%2Fi%3E%2C%20%3Ci%3E21%3C%5C%2Fi%3E%288%29%2C%20847%26%23x2013%3B857.%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Biomedical%20Hypothesis%20Generation%20by%20Text%20Mining%20and%20Gene%20Prioritization%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ingrid%22%2C%22lastName%22%3A%22Petri%5Cu010d%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bal%5Cu00e1zs%22%2C%22lastName%22%3A%22Ligeti%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bal%5Cu00e1zs%22%2C%22lastName%22%3A%22Gy%5Cu0151rffy%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%5D%2C%22abstractNote%22%3A%22Text%20mining%20methods%20can%20facilitate%20the%20generation%20of%20biomedical%20hypotheses%20by%20suggesting%20novel%20associations%20between%20diseases%20and%20genes.%20Previously%2C%20we%20developed%20a%20rare-term%20model%20called%20RaJoLink%20%28Petric%20et%20al%2C%20J.%20Biomed.%20Inform.%2042%282%29%3A%20219-227%2C%202009%29%20in%20which%20hypotheses%20are%20formulated%5Cnon%20the%20basis%20of%20terms%20rarely%20associated%20with%20a%20target%20domain.%20Since%20many%20current%20medical%20hypotheses%20are%20formulated%20in%20terms%20of%20molecular%20entities%20and%20molecular%20mechanisms%2C%20here%20we%20extend%20the%20methodology%20to%20proteins%20and%20genes%2C%20using%20a%20standardized%20vocabulary%20as%20well%20as%20a%20gene%5C%2Fprotein%20network%5Cnmodel.%20The%20proposed%20enhanced%20RaJoLink%20rare-term%20model%20combines%20text%20mining%20and%20gene%20prioritization%20approaches.%20Its%20utility%20is%20illustrated%20by%20finding%20known%20as%20well%20as%20potential%20gene-disease%20associations%20in%20ovarian%20cancer%20using%20MEDLINE%20abstracts%20and%20the%20STRING%20database.%22%2C%22date%22%3A%222014-06-01%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%22%22%2C%22ISSN%22%3A%22%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-10T12%3A06%3A43Z%22%7D%7D%2C%7B%22key%22%3A%22H4YVYQ6N%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Reiz%20and%20Pongor%22%2C%22parsedDate%22%3A%222011-09%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EReiz%2C%20B.%2C%20%26amp%3B%20Pongor%2C%20S.%20%282011%29.%20Psychologically%20Inspired%2C%20Rule-Based%20Outlier%20Detection%20in%20Noisy%20Data.%20%3Ci%3E13th%20International%20Symposium%20on%20Symbolic%20and%20Numeric%20Algorithms%20for%20Scientific%20Computing%3A%202011%3C%5C%2Fi%3E%2C%20%3Ci%3E1%3C%5C%2Fi%3E%2C%20131%26%23x2013%3B136.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1109%5C%2FSYNASC.2011.57%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1109%5C%2FSYNASC.2011.57%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22conferencePaper%22%2C%22title%22%3A%22Psychologically%20Inspired%2C%20Rule-Based%20Outlier%20Detection%20in%20Noisy%20Data%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Be%5Cu00e1ta%22%2C%22lastName%22%3A%22Reiz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%5D%2C%22abstractNote%22%3A%22Outlier%20detection%20is%20widely%20applied%20in%20several%20fields%20such%20as%20data%20mining%2C%20pattern%20recognition%20and%20bioinformatics.%20The%20algorithms%20used%20for%20outlier%20detection%20are%20based%20mainly%20on%20statistics%20and%20artificial%20intelligence.%20Our%20long-term%20goal%20is%20to%20study%20and%20apply%20the%20principles%20of%20human%20vision%20for%20solving%20outlier%20detection%20problems.%20Here%20we%20present%20an%20algorithm%20suitable%20for%20outlier%20detection%20based%20on%20the%20principles%20of%20Gestalt%20psychology.%20We%20demonstrate%20the%20algorithm%27s%20main%20properties%20on%20an%20example%20taken%20from%20human%20perception%2C%20the%20recognition%20of%20continuous%20curves%20formed%20of%20Gabor%20patches%20embedded%20into%20a%20noisy%20background.%20We%20show%20that%20the%20algorithm%20is%20tolerant%20with%20respect%20to%20added%20noise%20and%20is%20orientation%20independent.%20As%20a%20potential%20application%20we%20present%20the%20problem%20of%20filtering%20proteomics%20mass%20spectrometry%20data.%20The%20true%20peaks%20within%20a%20measured%20mass%20spectrum%20can%20be%20represented%20as%20a%20graph%20in%20which%20nodes%20are%20fragment%20peaks%20while%20edges%20represent%20equivalents%20of%20proximity%2C%20similarity%20and%20continuity%20defined%20in%20terms%20of%20chemical%20rules.%20The%20applicability%20of%20the%20principle%20to%20further%20problems%20is%20discussed.%22%2C%22date%22%3A%22Sep.%202011%22%2C%22proceedingsTitle%22%3A%2213th%20International%20Symposium%20on%20Symbolic%20and%20Numeric%20Algorithms%20for%20Scientific%20Computing%3A%202011%22%2C%22conferenceName%22%3A%222011%2013th%20International%20Symposium%20on%20Symbolic%20and%20Numeric%20Algorithms%20for%20Scientific%20Computing%22%2C%22language%22%3A%22%22%2C%22DOI%22%3A%2210.1109%5C%2FSYNASC.2011.57%22%2C%22ISBN%22%3A%22%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-06T20%3A54%3A33Z%22%7D%7D%2C%7B%22key%22%3A%22TDKLYCSX%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Csermely%20et%20al.%22%2C%22parsedDate%22%3A%222005-04%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ECsermely%2C%20P.%2C%20Agoston%2C%20V.%2C%20%26amp%3B%20Pongor%2C%20S.%20%282005%29.%20The%20efficiency%20of%20multi-target%20drugs%3A%20the%20network%20approach%20might%20help%20drug%20design.%20%3Ci%3ETrends%20in%20Pharmacological%20Sciences%3C%5C%2Fi%3E%2C%20%3Ci%3E26%3C%5C%2Fi%3E%284%29%2C%20178%26%23x2013%3B182.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tips.2005.02.007%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tips.2005.02.007%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22The%20efficiency%20of%20multi-target%20drugs%3A%20the%20network%20approach%20might%20help%20drug%20design%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22P%5Cu00e9ter%22%2C%22lastName%22%3A%22Csermely%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vilmos%22%2C%22lastName%22%3A%22Agoston%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%5D%2C%22abstractNote%22%3A%22Despite%20considerable%20progress%20in%20genome-%20and%20proteome-based%20high-throughput%20screening%20methods%20and%20rational%20drug%20design%2C%20the%20number%20of%20successful%20single-target%20drugs%20did%20not%20increase%20appreciably%20during%20the%20past%20decade.%20Network%20models%20suggest%20that%20partial%20inhibition%20of%20a%20surprisingly%20small%20number%20of%20targets%20can%20be%20more%20efficient%20than%20the%20complete%20inhibition%20of%20a%20single%20target.%20This%20and%20the%20success%20stories%20of%20multi-target%20drugs%20and%20combinatorial%20therapies%20led%20us%20to%20suggest%20that%20systematic%20drug-design%20strategies%20should%20be%20directed%20against%20multiple%20targets.%20We%20propose%20that%20the%20final%20effect%20of%20partial%2C%20but%20multiple%2C%20drug%20actions%20might%20often%20surpass%20that%20of%20complete%20drug%20action%20at%20a%20single%20target.%20The%20future%20success%20of%20this%20novel%20drug-design%20paradigm%20will%20depend%20not%20only%20on%20a%20new%20generation%20of%20computer%20models%20to%20identify%20the%20correct%20multiple%20targets%20and%20their%20multi-fitting%2C%20low-affinity%20drug%20candidates%20but%20also%20on%20more-efficient%20in%20vivo%20testing.%22%2C%22date%22%3A%22Apr%202005%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.tips.2005.02.007%22%2C%22ISSN%22%3A%220165-6147%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpubmed%5C%2F15808341%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-06T20%3A23%3A22Z%22%7D%7D%2C%7B%22key%22%3A%224VTZS4QB%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kert%5Cu00e9sz-Farkas%20et%20al.%22%2C%22parsedDate%22%3A%222011%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKert%26%23xE9%3Bsz-Farkas%2C%20A.%2C%20Reiz%2C%20B.%2C%20Myers%2C%20M.%20P.%2C%20%26amp%3B%20Pongor%2C%20S.%20%282011%29.%20PTMSearch%3A%20A%20Greedy%20Tree%20Traversal%20Algorithm%20for%20Finding%20Protein%20Post-Translational%20Modifications%20in%20Tandem%20Mass%20Spectra.%20In%20D.%20Gunopulos%2C%20T.%20Hofmann%2C%20D.%20Malerba%2C%20%26amp%3B%20M.%20Vazirgiannis%20%28Eds.%29%2C%20%3Ci%3EMachine%20Learning%20and%20Knowledge%20Discovery%20in%20Databases%3C%5C%2Fi%3E%20%28Vol.%202%2C%20pp.%20162%26%23x2013%3B176%29.%20Springer.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2F978-3-642-23783-6_11%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2F978-3-642-23783-6_11%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22conferencePaper%22%2C%22title%22%3A%22PTMSearch%3A%20A%20Greedy%20Tree%20Traversal%20Algorithm%20for%20Finding%20Protein%20Post-Translational%20Modifications%20in%20Tandem%20Mass%20Spectra%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Attila%22%2C%22lastName%22%3A%22Kert%5Cu00e9sz-Farkas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Be%5Cu00e1ta%22%2C%22lastName%22%3A%22Reiz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michael%20P.%22%2C%22lastName%22%3A%22Myers%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%2C%7B%22creatorType%22%3A%22editor%22%2C%22firstName%22%3A%22Dimitrios%22%2C%22lastName%22%3A%22Gunopulos%22%7D%2C%7B%22creatorType%22%3A%22editor%22%2C%22firstName%22%3A%22Thomas%22%2C%22lastName%22%3A%22Hofmann%22%7D%2C%7B%22creatorType%22%3A%22editor%22%2C%22firstName%22%3A%22Donato%22%2C%22lastName%22%3A%22Malerba%22%7D%2C%7B%22creatorType%22%3A%22editor%22%2C%22firstName%22%3A%22Michalis%22%2C%22lastName%22%3A%22Vazirgiannis%22%7D%5D%2C%22abstractNote%22%3A%22Peptide%20identification%20by%20tandem%20mass%20spectrometry%20%28MS%5C%2FMS%29%20and%20database%20searching%20is%20becoming%20the%20standard%20high-throughput%20technology%20in%20many%20areas%20of%20the%20life%20sciences.%20The%20analysis%20of%20post-translational%20modifications%20%28PTMs%29%20is%20a%20major%20source%20of%20complications%20in%20this%20area%2C%20which%20calls%20for%20efficient%20computational%20approaches.%20In%20this%20paper%20we%20describe%20PTMSearch%2C%20a%20novel%20algorithm%20in%20which%20the%20PTM%20search%20space%20is%20represented%20by%20a%20tree%20structure%2C%20and%20a%20greedy%20traversal%20algorithm%20is%20used%20to%20identify%20a%20path%20within%20the%20tree%20that%20corresponds%20to%20the%20PTMs%20that%20best%20fit%20the%20input%20data.%20Tests%20on%20simulated%20and%20real%20%28experimental%29%20PTMs%20show%20that%20the%20algorithm%20performs%20well%20in%20terms%20of%20speed%20and%20accuracy.%20Estimates%20are%20given%20for%20the%20error%20caused%20by%20the%20greedy%20heuristics%2C%20for%20the%20size%20of%20the%20search%20space%20and%20a%20scheme%20is%20presented%20for%20the%20calculation%20of%20statistical%20significance.%22%2C%22date%22%3A%222011%22%2C%22proceedingsTitle%22%3A%22Machine%20Learning%20and%20Knowledge%20Discovery%20in%20Databases%22%2C%22conferenceName%22%3A%22%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1007%5C%2F978-3-642-23783-6_11%22%2C%22ISBN%22%3A%22978-3-642-23783-6%22%2C%22url%22%3A%22%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-06T20%3A09%3A52Z%22%7D%7D%2C%7B%22key%22%3A%22VRCKVGQ9%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kuzniar%20et%20al.%22%2C%22parsedDate%22%3A%222009-07%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKuzniar%2C%20A.%2C%20Lin%2C%20K.%2C%20He%2C%20Y.%2C%20Nijveen%2C%20H.%2C%20Pongor%2C%20S.%2C%20%26amp%3B%20Leunissen%2C%20J.%20A.%20M.%20%282009%29.%20ProGMap%3A%20an%20integrated%20annotation%20resource%20for%20protein%20orthology.%20%3Ci%3ENucleic%20Acids%20Research%3C%5C%2Fi%3E%2C%20%3Ci%3E37%3C%5C%2Fi%3E%28Web%20Server%20issue%29%2C%20W428-434.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkp462%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fnar%5C%2Fgkp462%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22ProGMap%3A%20an%20integrated%20annotation%20resource%20for%20protein%20orthology%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Arnold%22%2C%22lastName%22%3A%22Kuzniar%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ke%22%2C%22lastName%22%3A%22Lin%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ying%22%2C%22lastName%22%3A%22He%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Harm%22%2C%22lastName%22%3A%22Nijveen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jack%20A.%20M.%22%2C%22lastName%22%3A%22Leunissen%22%7D%5D%2C%22abstractNote%22%3A%22Current%20protein%20sequence%20databases%20employ%20different%20classification%20schemes%20that%20often%20provide%20conflicting%20annotations%2C%20especially%20for%20poorly%20characterized%20proteins.%20ProGMap%20%28Protein%20Group%20Mappings%2C%20http%3A%5C%2F%5C%2Fwww.bioinformatics.nl%5C%2Fprogmap%29%20is%20a%20web-tool%20designed%20to%20help%20researchers%20and%20database%20annotators%20to%20assess%20the%20coherence%20of%20protein%20groups%20defined%20in%20various%20databases%20and%20thereby%20facilitate%20the%20annotation%20of%20newly%20sequenced%20proteins.%20ProGMap%20is%20based%20on%20a%20non-redundant%20dataset%20of%20over%206.6%20million%20protein%20sequences%20which%20is%20mapped%20to%20240%2C000%20protein%20group%20descriptions%20collected%20from%20UniProt%2C%20RefSeq%2C%20Ensembl%2C%20COG%2C%20KOG%2C%20OrthoMCL-DB%2C%20HomoloGene%2C%20TRIBES%20and%20PIRSF.%20ProGMap%20combines%20the%20underlying%20classification%20schemes%20via%20a%20network%20of%20links%20constructed%20by%20a%20fast%20and%20fully%20automated%20mapping%20approach%20originally%20developed%20for%20document%20classification.%20The%20web%20interface%20enables%20queries%20to%20be%20made%20using%20sequence%20identifiers%2C%20gene%20symbols%2C%20protein%20functions%20or%20amino%20acid%20and%20nucleotide%20sequences.%20For%20the%20latter%20query%20type%20BLAST%20similarity%20search%20and%20QuickMatch%20identity%20search%20services%20have%20been%20incorporated%2C%20for%20finding%20sequences%20similar%20%28or%20identical%29%20to%20a%20query%20sequence.%20ProGMap%20is%20meant%20to%20help%20users%20of%20high%20throughput%20methodologies%20who%20deal%20with%20partially%20annotated%20genomic%20data.%22%2C%22date%22%3A%22Jul%202009%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1093%5C%2Fnar%5C%2Fgkp462%22%2C%22ISSN%22%3A%221362-4962%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpubmed%5C%2F19494185%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-06T20%3A05%3A57Z%22%7D%7D%2C%7B%22key%22%3A%22YCIIGX2J%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kuzniar%20et%20al.%22%2C%22parsedDate%22%3A%222010-10-01%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKuzniar%2C%20A.%2C%20Dhir%2C%20S.%2C%20Nijveen%2C%20H.%2C%20Pongor%2C%20S.%2C%20%26amp%3B%20Leunissen%2C%20J.%20A.%20M.%20%282010%29.%20Multi-netclust%3A%20an%20efficient%20tool%20for%20finding%20connected%20clusters%20in%20multi-parametric%20networks.%20%3Ci%3EBioinformatics%20%28Oxford%2C%20England%29%3C%5C%2Fi%3E%2C%20%3Ci%3E26%3C%5C%2Fi%3E%2819%29%2C%202482%26%23x2013%3B2483.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fbioinformatics%5C%2Fbtq435%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fbioinformatics%5C%2Fbtq435%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Multi-netclust%3A%20an%20efficient%20tool%20for%20finding%20connected%20clusters%20in%20multi-parametric%20networks%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Arnold%22%2C%22lastName%22%3A%22Kuzniar%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Somdutta%22%2C%22lastName%22%3A%22Dhir%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Harm%22%2C%22lastName%22%3A%22Nijveen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jack%20A.%20M.%22%2C%22lastName%22%3A%22Leunissen%22%7D%5D%2C%22abstractNote%22%3A%22Multi-netclust%20is%20a%20simple%20tool%20that%20allows%20users%20to%20extract%20connected%20clusters%20of%20data%20represented%20by%20different%20networks%20given%20in%20the%20form%20of%20matrices.%20The%20tool%20uses%20user-defined%20threshold%20values%20to%20combine%20the%20matrices%2C%20and%20uses%20a%20straightforward%2C%20memory-efficient%20graph%20algorithm%20to%20find%20clusters%20that%20are%20connected%20in%20all%20or%20in%20either%20of%20the%20networks.%20The%20tool%20is%20written%20in%20C%5C%2FC%2B%2B%20and%20is%20available%20either%20as%20a%20form-based%20or%20as%20a%20command-line-based%20program%20running%20on%20Linux%20platforms.%20The%20algorithm%20is%20fast%2C%20processing%20a%20network%20of%20%3E%2010%286%29%20nodes%20and%2010%288%29%20edges%20takes%20only%20a%20few%20minutes%20on%20an%20ordinary%20computer.%5CnAVAILABILITY%3A%20http%3A%5C%2F%5C%2Fwww.bioinformatics.nl%5C%2Fnetclust%5C%2F.%22%2C%22date%22%3A%22Oct%2001%2C%202010%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1093%5C%2Fbioinformatics%5C%2Fbtq435%22%2C%22ISSN%22%3A%221367-4811%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpubmed%5C%2F20679333%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-06T19%3A41%3A51Z%22%7D%7D%2C%7B%22key%22%3A%22ZMJY9KY8%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Vera%20et%20al.%22%2C%22parsedDate%22%3A%222013-01-01%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EVera%2C%20R.%2C%20Perez-Riverol%2C%20Y.%2C%20Perez%2C%20S.%2C%20Ligeti%2C%20B.%2C%20Kert%26%23xE9%3Bsz-Farkas%2C%20A.%2C%20%26amp%3B%20Pongor%2C%20S.%20%282013%29.%20JBioWH%3A%20an%20open-source%20Java%20framework%20for%20bioinformatics%20data%20integration.%20%3Ci%3EDatabase%3C%5C%2Fi%3E%2C%20%3Ci%3E2013%3C%5C%2Fi%3E%2C%20bat051.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fdatabase%5C%2Fbat051%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fdatabase%5C%2Fbat051%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22JBioWH%3A%20an%20open-source%20Java%20framework%20for%20bioinformatics%20data%20integration%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Roberto%22%2C%22lastName%22%3A%22Vera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yasset%22%2C%22lastName%22%3A%22Perez-Riverol%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sonia%22%2C%22lastName%22%3A%22Perez%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bal%5Cu00e1zs%22%2C%22lastName%22%3A%22Ligeti%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Attila%22%2C%22lastName%22%3A%22Kert%5Cu00e9sz-Farkas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%5D%2C%22abstractNote%22%3A%22Abstract.%20%20The%20Java%20BioWareHouse%20%28JBioWH%29%20project%20is%20an%20open-source%20platform-independent%20programming%20framework%20that%20allows%20a%20user%20to%20build%20his%5C%2Fher%20own%20integrate%22%2C%22date%22%3A%222013%5C%2F01%5C%2F01%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1093%5C%2Fdatabase%5C%2Fbat051%22%2C%22ISSN%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Facademic.oup.com%5C%2Fdatabase%5C%2Farticle%5C%2Fdoi%5C%2F10.1093%5C%2Fdatabase%5C%2Fbat051%5C%2F337984%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-06T19%3A40%3A34Z%22%7D%7D%2C%7B%22key%22%3A%22SN743IDY%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Reiz%20et%20al.%22%2C%22parsedDate%22%3A%222013-06-19%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EReiz%2C%20B.%2C%20Busa-Fekete%2C%20R.%2C%20Pongor%2C%20S.%2C%20%26amp%3B%20Kov%26%23xE1%3Bcs%2C%20I.%20%282013%29.%20Closure%20enhancement%20in%20a%20model%20network%20with%20orientation%20tuned%20long-range%20connectivity.%20%3Ci%3ELearning%20%26amp%3B%20Perception%3C%5C%2Fi%3E%2C%20%3Ci%3E5%3C%5C%2Fi%3E%28Supplement-2%29%2C%20119%26%23x2013%3B148.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1556%5C%2Flp.5.2013.suppl2.8%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1556%5C%2Flp.5.2013.suppl2.8%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Closure%20enhancement%20in%20a%20model%20network%20with%20orientation%20tuned%20long-range%20connectivity%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Be%5Cu00e1ta%22%2C%22lastName%22%3A%22Reiz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R%5Cu00f3bert%22%2C%22lastName%22%3A%22Busa-Fekete%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Ilona%22%2C%22lastName%22%3A%22Kov%5Cu00e1cs%22%7D%5D%2C%22abstractNote%22%3A%22%3Csection%20class%3D%5C%22abstract%5C%22%3E%3Cp%3EThe%20primary%20visual%20cortex%20%28V1%29%20of%20the%20mammalian%20brain%20is%20equipped%20with%20a%20specifically%20connected%20network%20of%20neurons%20that%20can%20potentially%20solve%20difficult%20image%20processing%20tasks.%20These%20neurons%20are%20selectively%20tuned%20for%20locations%20in%20visual%20space%20and%20also%20for%20line%20orientation.%20The%20coupling%20of%20location%20and%20orientation%20tuning%20results%20in%20the%20neural%20representation%20of%20the%20visual%20world%20in%20terms%20of%20local%20features.%20These%20local%20features%2C%20e.g.%2C%20oriented%20line%20segments%2C%20will%20have%20to%20be%20linked%20together%20in%20order%20to%20parse%20the%20visual%20world%20into%20regions%20corresponding%20to%20object%20and%20ground.%20Although%20standard%20models%20of%20V1%20do%20not%20address%20the%20issue%20of%20interacting%20neuronal%20populations%2C%20we%20suggest%20that%20the%20long-range%20connectivity%20pattern%20of%20V1%20provides%20an%20architecture%20where%20spreading%20neural%20activity%20may%20lead%20to%20pertinent%20figure-ground%20segmentation.%20The%20model%20relies%20on%20the%20fact%20that%20in%20addition%20to%20the%20processing%20units%2C%20their%20connections%20are%20also%20selectively%20tuned%20for%20space%20and%20orientation.%20From%20the%20computational%20point%20of%20view%2C%20the%20model%20uses%20a%20minimalist%20approach%20that%20applies%20the%20fundamental%20concepts%20of%20Gestalt%20psychology%20%5Cu2013%20proximity%2C%20similarity%20and%20continuity%20%5Cu2013%20to%20the%20spreading%20of%20neuronal%20activation%20signals.%20This%20model%20is%20successful%20in%20predicting%20psychophysical%20performance%20of%20human%20observers%2C%20and%20provides%20an%20account%20of%20the%20computational%20power%20of%20V1.%3C%5C%2Fp%3E%3C%5C%2Fsection%3E%22%2C%22date%22%3A%222013%5C%2F06%5C%2F19%22%2C%22language%22%3A%22en_US%22%2C%22DOI%22%3A%2210.1556%5C%2Flp.5.2013.suppl2.8%22%2C%22ISSN%22%3A%222060-9175%2C%201789-3186%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fakjournals.com%5C%2Fview%5C%2Fjournals%5C%2F1166%5C%2F5%5C%2FSupplement-2%5C%2Farticle-p119.xml%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-06T19%3A17%3A19Z%22%7D%7D%2C%7B%22key%22%3A%22ITV6C94W%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Reiz%20et%20al.%22%2C%22parsedDate%22%3A%222013-04-01%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EReiz%2C%20B.%2C%20Kert%26%23xE9%3Bsz-Farkas%2C%20A.%2C%20Pongor%2C%20S.%2C%20%26amp%3B%20Myers%2C%20M.%20P.%20%282013%29.%20Chemical%20rule-based%20filtering%20of%20MS%5C%2FMS%20spectra.%20%3Ci%3EBioinformatics%3C%5C%2Fi%3E%2C%20%3Ci%3E29%3C%5C%2Fi%3E%287%29%2C%20925%26%23x2013%3B932.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fbioinformatics%5C%2Fbtt061%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fbioinformatics%5C%2Fbtt061%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Chemical%20rule-based%20filtering%20of%20MS%5C%2FMS%20spectra%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Be%5Cu00e1ta%22%2C%22lastName%22%3A%22Reiz%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Attila%22%2C%22lastName%22%3A%22Kert%5Cu00e9sz-Farkas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Michael%20P.%22%2C%22lastName%22%3A%22Myers%22%7D%5D%2C%22abstractNote%22%3A%22Abstract.%20%20Motivation%3A%20Identification%20of%20proteins%20by%20mass%20spectrometry%5Cu2013based%20proteomics%20requires%20automated%20interpretation%20of%20peptide%20tandem%20mass%20spectrometry%20sp%22%2C%22date%22%3A%222013%5C%2F04%5C%2F01%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1093%5C%2Fbioinformatics%5C%2Fbtt061%22%2C%22ISSN%22%3A%221367-4803%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Facademic.oup.com%5C%2Fbioinformatics%5C%2Farticle%5C%2F29%5C%2F7%5C%2F925%5C%2F253395%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-06T19%3A12%3A50Z%22%7D%7D%5D%7DCserháti, M., Turóczy, Z., Zombori, Z., Cserzö, M., Dudits, D., Pongor, S., & Györgyey, J. (2011). Prediction of new abiotic stress genes in Arabidopsis thaliana and Oryza sativa according to enumeration-based statistical analysis.
Molecular Genetics and Genomics: MGG,
285(5), 375–391.
https://doi.org/10.1007/s00438-011-0605-4Reiz, B., P. Myers, M., Pongor, S., & Kertész-Farkas, A. (2014). Precursor Mass Dependent Filtering of Mass Spectra for Proteomics Analysis. Protein and Peptide Letters, 21(8), 858–863.
Kertész-Farkas, A., Reiz, B., Myers, M. P., & Pongor, S. (2012). Database Searching In Mass Spectrometry Based Proteomics. Current Bioinformatics, 7(2), 221–230.
Reiz, B., Kertész-Farkas, A., Pongor, S., & Myers, M. P. (2012). Data Preprocessing and Filtering in Mass Spectrometry Based Proteomics.
Current Bioinformatics,
7(2), 212–220.
https://doi.org/10.2174/157489312800604363Petrič, I., Ligeti, B., Győrffy, B., & Pongor, S. (2014). Biomedical Hypothesis Generation by Text Mining and Gene Prioritization. Protein and Peptide Letters, 21(8), 847–857.
Reiz, B., & Pongor, S. (2011). Psychologically Inspired, Rule-Based Outlier Detection in Noisy Data.
13th International Symposium on Symbolic and Numeric Algorithms for Scientific Computing: 2011,
1, 131–136.
https://doi.org/10.1109/SYNASC.2011.57Csermely, P., Agoston, V., & Pongor, S. (2005). The efficiency of multi-target drugs: the network approach might help drug design.
Trends in Pharmacological Sciences,
26(4), 178–182.
https://doi.org/10.1016/j.tips.2005.02.007Kertész-Farkas, A., Reiz, B., Myers, M. P., & Pongor, S. (2011). PTMSearch: A Greedy Tree Traversal Algorithm for Finding Protein Post-Translational Modifications in Tandem Mass Spectra. In D. Gunopulos, T. Hofmann, D. Malerba, & M. Vazirgiannis (Eds.),
Machine Learning and Knowledge Discovery in Databases (Vol. 2, pp. 162–176). Springer.
https://doi.org/10.1007/978-3-642-23783-6_11Kuzniar, A., Lin, K., He, Y., Nijveen, H., Pongor, S., & Leunissen, J. A. M. (2009). ProGMap: an integrated annotation resource for protein orthology.
Nucleic Acids Research,
37(Web Server issue), W428-434.
https://doi.org/10.1093/nar/gkp462Kuzniar, A., Dhir, S., Nijveen, H., Pongor, S., & Leunissen, J. A. M. (2010). Multi-netclust: an efficient tool for finding connected clusters in multi-parametric networks.
Bioinformatics (Oxford, England),
26(19), 2482–2483.
https://doi.org/10.1093/bioinformatics/btq435Vera, R., Perez-Riverol, Y., Perez, S., Ligeti, B., Kertész-Farkas, A., & Pongor, S. (2013). JBioWH: an open-source Java framework for bioinformatics data integration.
Database,
2013, bat051.
https://doi.org/10.1093/database/bat051Reiz, B., Busa-Fekete, R., Pongor, S., & Kovács, I. (2013). Closure enhancement in a model network with orientation tuned long-range connectivity.
Learning & Perception,
5(Supplement-2), 119–148.
https://doi.org/10.1556/lp.5.2013.suppl2.8Reiz, B., Kertész-Farkas, A., Pongor, S., & Myers, M. P. (2013). Chemical rule-based filtering of MS/MS spectra.
Bioinformatics,
29(7), 925–932.
https://doi.org/10.1093/bioinformatics/btt061