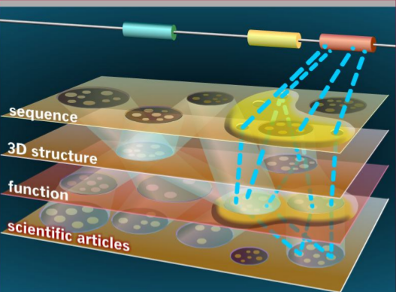
While traditional bioinformatics has evolved from simple data management to data-interpretation, the emphasis today has shifted to high-throughput data collection, personal medicine, and the analysis of complex systems. This tendency is accompanied by an unprecedented development of new computer architectures and cloud computing that bring the power of supercomputers within arm’s reach of bench scientists and clinical practitioners. At the same time, bionic devices and on-line diagnostic tools open up new areas of applications.
In this fast-evolving scene of new technologies, integrating heterogeneous bioinformatics data is perhaps one of the most challenging tasks. Databases increase both in volume and in complexity, and public resources available on the Internet can not cope with a growing number of user groups, especially medical and industrial users concerned with data confidentiality. On the other hand many, if not most biomolecular mechanisms that translate the human genomic information into phenotypes are not known and as a consequence, most of the molecular and cellular data cannot be interpreted in terms of biomedically relevant conclusions. While personalized diagnostics and cures are likely to remain a dominant trend, the temperate view suggests biomedical applications relying on the comparison of biomolecular sequences and/or on the already known biomolecular mechanisms may have even greater chances to enter clinical practice. Developing stand-alone tools for genome annotation, personalized medicine and high throughput technologies is especially important in the analysis of complex diseases such as neurological and psychiatric disorders.
Project Participants: Zsolt Gelencsér, PhD student Áron Erdei, BSc student Prof. Sándor Pongor, PI Collaborators Prof. Mária Judit Molnár Prof. Frank Eisenhaber Dr. Michael P. Myers Roberto Vera References 2542344
{2542344:JL9JZWQ5},{2542344:ZMJY9KY8},{2542344:TKZ2KWQ6},{2542344:2XQIYEY3},{2542344:99K2G9RY},{2542344:W2P99XAZ},{2542344:WD4RP6I9},{2542344:HDI33P33}
items
1
apa
0
default
asc
184
https://pongor2.itk.ppke.hu/wp-content/plugins/zotpress/
%7B%22status%22%3A%22success%22%2C%22updateneeded%22%3Afalse%2C%22instance%22%3A%22zotpress-9396c755c560e5ec6efdc1907d5a1bad%22%2C%22meta%22%3A%7B%22request_last%22%3A0%2C%22request_next%22%3A0%2C%22used_cache%22%3Atrue%7D%2C%22data%22%3A%5B%7B%22key%22%3A%2299K2G9RY%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kert%5Cu00e9sz-Farkas%20et%20al.%22%2C%22parsedDate%22%3A%222009%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKert%26%23xE9%3Bsz-Farkas%2C%20A.%2C%20Kocsor%2C%20A.%2C%20%26amp%3B%20Pongor%2C%20S.%20%282009%29.%20The%20Application%20of%20Data%20Compression-Based%20Distances%20to%20Biological%20Sequences.%20In%20F.%20Emmert-Streib%20%26amp%3B%20M.%20Dehmer%20%28Eds.%29%2C%20%3Ci%3EInformation%20Theory%20and%20Statistical%20Learning%3C%5C%2Fi%3E%20%28pp.%2083%26%23x2013%3B100%29.%20Springer%20US.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2F978-0-387-84816-7_4%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2F978-0-387-84816-7_4%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22bookSection%22%2C%22title%22%3A%22The%20Application%20of%20Data%20Compression-Based%20Distances%20to%20Biological%20Sequences%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Attila%22%2C%22lastName%22%3A%22Kert%5Cu00e9sz-Farkas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Andr%5Cu00e1s%22%2C%22lastName%22%3A%22Kocsor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%2C%7B%22creatorType%22%3A%22editor%22%2C%22firstName%22%3A%22Frank%22%2C%22lastName%22%3A%22Emmert-Streib%22%7D%2C%7B%22creatorType%22%3A%22editor%22%2C%22firstName%22%3A%22Matthias%22%2C%22lastName%22%3A%22Dehmer%22%7D%5D%2C%22abstractNote%22%3A%22Text%20compressor%20algorithms%20can%20be%20used%20to%20construct%20metric%20distance%20measures%20%28CBDs%29%20suitable%20for%20character%20sequences.%20Here%20we%20review%20the%20principle%20of%20various%20types%20of%20compressor%20algorithms%20and%20describe%20their%20general%20behaviour%20with%20respect%20to%20the%20comparison%20of%20protein%20and%20DNA%20sequences.%20We%20employ%20reduced%20and%20enlarged%20alphabets%2C%20and%20model%20biological%20rearrangements%20like%20domain%20shuffling.%20In%20the%20classification%20experiments%20evaluated%20with%20ROC%20analysis%2C%20CBDs%20perform%20less%20well%20than%20substring-based%20methods%20such%20as%20the%20BLAST%20and%20the%20Smith%5Cu2014Waterman%20algorithms%2C%20but%20perform%20better%20than%20distances%20based%20on%20word%20composition.%20CBDs%20outperformed%20substring%20methods%20with%20respect%20to%20domain%20shuffling%2C%20and%20in%20some%20cases%20showed%20an%20increased%20performance%20when%20the%20alphabet%20was%20reduced.%22%2C%22bookTitle%22%3A%22Information%20Theory%20and%20Statistical%20Learning%22%2C%22date%22%3A%222009%22%2C%22language%22%3A%22en%22%2C%22ISBN%22%3A%22978-0-387-84816-7%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1007%5C%2F978-0-387-84816-7_4%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-20T22%3A48%3A03Z%22%7D%7D%2C%7B%22key%22%3A%22JL9JZWQ5%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kuznetsov%20et%20al.%22%2C%22parsedDate%22%3A%222013-01-10%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKuznetsov%2C%20V.%2C%20Lee%2C%20H.%20K.%2C%20Maurer-Stroh%2C%20S.%2C%20Moln%26%23xE1%3Br%2C%20M.%20J.%2C%20Pongor%2C%20S.%2C%20Eisenhaber%2C%20B.%2C%20%26amp%3B%20Eisenhaber%2C%20F.%20%282013%29.%20How%20bioinformatics%20influences%20health%20informatics%3A%20usage%20of%20biomolecular%20sequences%2C%20expression%20profiles%20and%20automated%20microscopic%20image%20analyses%20for%20clinical%20needs%20and%20public%20health.%20%3Ci%3EHealth%20Information%20Science%20and%20Systems%3C%5C%2Fi%3E%2C%20%3Ci%3E1%3C%5C%2Fi%3E%281%29%2C%202.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1186%5C%2F2047-2501-1-2%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1186%5C%2F2047-2501-1-2%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22How%20bioinformatics%20influences%20health%20informatics%3A%20usage%20of%20biomolecular%20sequences%2C%20expression%20profiles%20and%20automated%20microscopic%20image%20analyses%20for%20clinical%20needs%20and%20public%20health%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Vladimir%22%2C%22lastName%22%3A%22Kuznetsov%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Hwee%20Kuan%22%2C%22lastName%22%3A%22Lee%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sebastian%22%2C%22lastName%22%3A%22Maurer-Stroh%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Maria%20Judit%22%2C%22lastName%22%3A%22Moln%5Cu00e1r%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Birgit%22%2C%22lastName%22%3A%22Eisenhaber%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Frank%22%2C%22lastName%22%3A%22Eisenhaber%22%7D%5D%2C%22abstractNote%22%3A%22The%20currently%20hyped%20expectation%20of%20personalized%20medicine%20is%20often%20associated%20with%20just%20achieving%20the%20information%20technology%20led%20integration%20of%20biomolecular%20sequencing%2C%20expression%20and%20histopathological%20bioimaging%20data%20with%20clinical%20records%20at%20the%20individual%20patients%5Cu2019%20level%20as%20if%20the%20significant%20biomedical%20conclusions%20would%20be%20its%20more%20or%20less%20mandatory%20result.%20It%20remains%20a%20sad%20fact%20that%20many%2C%20if%20not%20most%20biomolecular%20mechanisms%20that%20translate%20the%20human%20genomic%20information%20into%20phenotypes%20are%20not%20known%20and%2C%20thus%2C%20most%20of%20the%20molecular%20and%20cellular%20data%20cannot%20be%20interpreted%20in%20terms%20of%20biomedically%20relevant%20conclusions.%20Whereas%20the%20historical%20trend%20will%20certainly%20be%20into%20the%20general%20direction%20of%20personalized%20diagnostics%20and%20cures%2C%20the%20temperate%20view%20suggests%20that%20biomedical%20applications%20that%20rely%20either%20on%20the%20comparison%20of%20biomolecular%20sequences%20and%5C%2For%20on%20the%20already%20known%20biomolecular%20mechanisms%20have%20much%20greater%20chances%20to%20enter%20clinical%20practice%20soon.%20In%20addition%20to%20considering%20the%20general%20trends%2C%20we%20exemplarily%20review%20advances%20in%20the%20area%20of%20cancer%20biomarker%20discovery%2C%20in%20the%20clinically%20relevant%20characterization%20of%20patient-specific%20viral%20and%20bacterial%20pathogens%20%28with%20emphasis%20on%20drug%20selection%20for%20influenza%20and%20enterohemorrhagic%20E.%20coli%29%20as%20well%20as%20progress%20in%20the%20automated%20assessment%20of%20histopathological%20images.%20As%20molecular%20and%20cellular%20data%20analysis%20will%20become%20instrumental%20for%20achieving%20desirable%20clinical%20outcomes%2C%20the%20role%20of%20bioinformatics%20and%20computational%20biology%20approaches%20will%20dramatically%20grow.%22%2C%22date%22%3A%222013-01-10%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1186%5C%2F2047-2501-1-2%22%2C%22ISSN%22%3A%222047-2501%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1186%5C%2F2047-2501-1-2%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-10T12%3A39%3A56Z%22%7D%7D%2C%7B%22key%22%3A%22TKZ2KWQ6%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Busa-Fekete%20et%20al.%22%2C%22parsedDate%22%3A%222008%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EBusa-Fekete%2C%20R.%2C%20Kert%26%23xE9%3Bsz-Farkas%2C%20A.%2C%20Kocsor%2C%20A.%2C%20%26amp%3B%20Pongor%2C%20S.%20%282008%29.%20Balanced%20ROC%20%28BaROC%29%20analysis%20for%20portien%20classification.%20%3Ci%3EJournal%20of%20Biochemical%20and%20Biophysical%20Methods%3C%5C%2Fi%3E%2C%20%3Ci%3E70%3C%5C%2Fi%3E%286%29%2C%201210%26%23x2013%3B1214.%20%3Ca%20href%3D%27http%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpubmed%5C%2F17689617%27%3Ehttp%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpubmed%5C%2F17689617%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Balanced%20ROC%20%28BaROC%29%20analysis%20for%20portien%20classification%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R%5Cu00f3bert%22%2C%22lastName%22%3A%22Busa-Fekete%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Attila%22%2C%22lastName%22%3A%22Kert%5Cu00e9sz-Farkas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Andr%5Cu00e1s%22%2C%22lastName%22%3A%22Kocsor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%5D%2C%22abstractNote%22%3A%22Identification%20of%20problematic%20protein%20classes%20%28domain%20types%2C%20protein%20families%29%20that%20are%20difficult%20to%20predict%20from%20sequence%20is%20a%20key%20issue%20in%20genome%20annotation.%20ROC%20%28Receiver%20Operating%20Characteristic%29%20analysis%20is%20routinely%20used%20for%20the%20evaluation%20of%20protein%20similarities%2C%20however%20its%20results%20%3F%20the%20area%20under%20curve%20%28AUC%29%20values%20%3F%20are%20differentially%20biased%20for%20the%20various%20protein%20classes%20that%20are%20highly%20different%20in%20size.%20We%20show%20the%20bias%20can%20be%20compensated%20for%20by%20adjusting%20the%20length%20of%20the%20top%20list%20in%20a%20class-dependent%20fashion%2C%20so%20that%20the%20number%20of%20negatives%20within%20the%20top%20list%20will%20be%20equal%20to%20%28or%20proportional%20with%29%20the%20size%20of%20the%20positive%20class.%20Using%20this%20balanced%20protocol%20the%20problematic%20classes%20can%20be%20identified%20by%20their%20AUC%20values%2C%20or%20by%20a%20scatter%20diagram%20in%20which%20the%20AUC%20values%20are%20plotted%20against%20positive%5C%2Fnegative%20ratio%20of%20the%20top%20list.%20The%20use%20of%20likelihood-ratio%20scoring%20%28Kajan%20et%20al%2C%20Bioinformatics%2C22%2C%202865-2869%2C%202007%29%20the%20bias%20caused%20by%20class%20imbalance%20can%20be%20further%20decreased.%22%2C%22date%22%3A%222008%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%22%22%2C%22ISSN%22%3A%22%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpubmed%5C%2F17689617%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-10T11%3A36%3A55Z%22%7D%7D%2C%7B%22key%22%3A%22WD4RP6I9%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kuzniar%20et%20al.%22%2C%22parsedDate%22%3A%222008-11%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKuzniar%2C%20A.%2C%20van%20Ham%2C%20R.%20C.%20H.%20J.%2C%20Pongor%2C%20S.%2C%20%26amp%3B%20Leunissen%2C%20J.%20A.%20M.%20%282008%29.%20The%20quest%20for%20orthologs%3A%20finding%20the%20corresponding%20gene%20across%20genomes.%20%3Ci%3ETrends%20in%20Genetics%3A%20TIG%3C%5C%2Fi%3E%2C%20%3Ci%3E24%3C%5C%2Fi%3E%2811%29%2C%20539%26%23x2013%3B551.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tig.2008.08.009%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.tig.2008.08.009%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22The%20quest%20for%20orthologs%3A%20finding%20the%20corresponding%20gene%20across%20genomes%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Arnold%22%2C%22lastName%22%3A%22Kuzniar%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Roeland%20C.%20H.%20J.%22%2C%22lastName%22%3A%22van%20Ham%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jack%20A.%20M.%22%2C%22lastName%22%3A%22Leunissen%22%7D%5D%2C%22abstractNote%22%3A%22Orthology%20is%20a%20key%20evolutionary%20concept%20in%20many%20areas%20of%20genomic%20research.%20It%20provides%20a%20framework%20for%20subjects%20as%20diverse%20as%20the%20evolution%20of%20genomes%2C%20gene%20functions%2C%20cellular%20networks%20and%20functional%20genome%20annotation.%20Although%20orthologous%20proteins%20usually%20perform%20equivalent%20functions%20in%20different%20species%2C%20establishing%20true%20orthologous%20relationships%20requires%20a%20phylogenetic%20approach%2C%20which%20combines%20both%20trees%20and%20graphs%20%28networks%29%20using%20reliable%20species%20phylogeny%20and%20available%20genomic%20data%20from%20more%20than%20two%20species%2C%20and%20an%20insight%20into%20the%20processes%20of%20molecular%20evolution.%20Here%2C%20we%20evaluate%20the%20available%20bioinformatics%20tools%20and%20provide%20a%20set%20of%20guidelines%20to%20aid%20researchers%20in%20choosing%20the%20most%20appropriate%20tool%20for%20any%20situation.%22%2C%22date%22%3A%22Nov%202008%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.tig.2008.08.009%22%2C%22ISSN%22%3A%220168-9525%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpubmed%5C%2F18819722%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-10T10%3A49%3A33Z%22%7D%7D%2C%7B%22key%22%3A%222XQIYEY3%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kert%5Cu00e9sz-Farkas%20et%20al.%22%2C%22parsedDate%22%3A%222008-04-24%22%2C%22numChildren%22%3A4%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKert%26%23xE9%3Bsz-Farkas%2C%20A.%2C%20Dhir%2C%20S.%2C%20Sonego%2C%20P.%2C%20Pacurar%2C%20M.%2C%20Netoteia%2C%20S.%2C%20Nijveen%2C%20H.%2C%20Kuzniar%2C%20A.%2C%20Leunissen%2C%20J.%20A.%20M.%2C%20Kocsor%2C%20A.%2C%20%26amp%3B%20Pongor%2C%20S.%20%282008%29.%20Benchmarking%20protein%20classification%20algorithms%20via%20supervised%20cross-validation.%20%3Ci%3EJournal%20of%20Biochemical%20and%20Biophysical%20Methods%3C%5C%2Fi%3E%2C%20%3Ci%3E70%3C%5C%2Fi%3E%286%29%2C%201215%26%23x2013%3B1223.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.jbbm.2007.05.011%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1016%5C%2Fj.jbbm.2007.05.011%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Benchmarking%20protein%20classification%20algorithms%20via%20supervised%20cross-validation%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Attila%22%2C%22lastName%22%3A%22Kert%5Cu00e9sz-Farkas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Somdutta%22%2C%22lastName%22%3A%22Dhir%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Paolo%22%2C%22lastName%22%3A%22Sonego%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Mircea%22%2C%22lastName%22%3A%22Pacurar%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sergiu%22%2C%22lastName%22%3A%22Netoteia%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Harm%22%2C%22lastName%22%3A%22Nijveen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Arnold%22%2C%22lastName%22%3A%22Kuzniar%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Jack%20A.%20M.%22%2C%22lastName%22%3A%22Leunissen%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Andr%5Cu00e1s%22%2C%22lastName%22%3A%22Kocsor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%5D%2C%22abstractNote%22%3A%22Development%20and%20testing%20of%20protein%20classification%20algorithms%20are%20hampered%20by%20the%20fact%20that%20the%20protein%20universe%20is%20characterized%20by%20groups%20vastly%20different%20in%20the%20number%20of%20members%2C%20in%20average%20protein%20size%2C%20similarity%20within%20group%2C%20etc.%20Datasets%20based%20on%20traditional%20cross-validation%20%28k-fold%2C%20leave-one-out%2C%20etc.%29%20may%20not%20give%20reliable%20estimates%20on%20how%20an%20algorithm%20will%20generalize%20to%20novel%2C%20distantly%20related%20subtypes%20of%20the%20known%20protein%20classes.%20Supervised%20cross-validation%2C%20i.e.%2C%20selection%20of%20test%20and%20train%20sets%20according%20to%20the%20known%20subtypes%20within%20a%20database%20has%20been%20successfully%20used%20earlier%20in%20conjunction%20with%20the%20SCOP%20database.%20Our%20goal%20was%20to%20extend%20this%20principle%20to%20other%20databases%20and%20to%20design%20standardized%20benchmark%20datasets%20for%20protein%20classification.%20Hierarchical%20classification%20trees%20of%20protein%20categories%20provide%20a%20simple%20and%20general%20framework%20for%20designing%20supervised%20cross-validation%20strategies%20for%20protein%20classification.%20Benchmark%20datasets%20can%20be%20designed%20at%20various%20levels%20of%20the%20concept%20hierarchy%20using%20a%20simple%20graph-theoretic%20distance.%20A%20combination%20of%20supervised%20and%20random%20sampling%20was%20selected%20to%20construct%20reduced%20size%20model%20datasets%2C%20suitable%20for%20algorithm%20comparison.%20Over%203000%20new%20classification%20tasks%20were%20added%20to%20our%20recently%20established%20protein%20classification%20benchmark%20collection%20that%20currently%20includes%20protein%20sequence%20%28including%20protein%20domains%20and%20entire%20proteins%29%2C%20protein%20structure%20and%20reading%20frame%20DNA%20sequence%20data.%20We%20carried%20out%20an%20extensive%20evaluation%20based%20on%20various%20machine-learning%20algorithms%20such%20as%20nearest%20neighbor%2C%20support%20vector%20machines%2C%20artificial%20neural%20networks%2C%20random%20forests%20and%20logistic%20regression%2C%20used%20in%20conjunction%20with%20comparison%20algorithms%2C%20BLAST%2C%20Smith-Waterman%2C%20Needleman-Wunsch%2C%20as%20well%20as%203D%20comparison%20methods%20DALI%20and%20PRIDE.%20The%20resulting%20datasets%20provide%20lower%2C%20and%20in%20our%20opinion%20more%20realistic%20estimates%20of%20the%20classifier%20performance%20than%20do%20random%20cross-validation%20schemes.%20A%20combination%20of%20supervised%20and%20random%20sampling%20was%20used%20to%20construct%20model%20datasets%2C%20suitable%20for%20algorithm%20comparison.%22%2C%22date%22%3A%22Apr%2024%2C%202008%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1016%5C%2Fj.jbbm.2007.05.011%22%2C%22ISSN%22%3A%220165-022X%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpubmed%5C%2F17604112%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-10T10%3A43%3A52Z%22%7D%7D%2C%7B%22key%22%3A%22HDI33P33%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Sonego%20et%20al.%22%2C%22parsedDate%22%3A%222008-05%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3ESonego%2C%20P.%2C%20Kocsor%2C%20A.%2C%20%26amp%3B%20Pongor%2C%20S.%20%282008%29.%20ROC%20analysis%3A%20applications%20to%20the%20classification%20of%20biological%20sequences%20and%203D%20structures.%20%3Ci%3EBriefings%20in%20Bioinformatics%3C%5C%2Fi%3E%2C%20%3Ci%3E9%3C%5C%2Fi%3E%283%29%2C%20198%26%23x2013%3B209.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fbib%5C%2Fbbm064%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fbib%5C%2Fbbm064%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22ROC%20analysis%3A%20applications%20to%20the%20classification%20of%20biological%20sequences%20and%203D%20structures%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Paolo%22%2C%22lastName%22%3A%22Sonego%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Andr%5Cu00e1s%22%2C%22lastName%22%3A%22Kocsor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%5D%2C%22abstractNote%22%3A%22ROC%20%28%27receiver%20operator%20characteristics%27%29%20analysis%20is%20a%20visual%20as%20well%20as%20numerical%20method%20used%20for%20assessing%20the%20performance%20of%20classification%20algorithms%2C%20such%20as%20those%20used%20for%20predicting%20structures%20and%20functions%20from%20sequence%20data.%20This%20review%20summarizes%20the%20fundamental%20concepts%20of%20ROC%20analysis%20and%20the%20interpretation%20of%20results%20using%20examples%20of%20sequence%20and%20structure%20comparison.%20We%20overview%20the%20available%20programs%20and%20provide%20evaluation%20guidelines%20for%20genomic%5C%2Fproteomic%20data%2C%20with%20particular%20regard%20to%20applications%20to%20large%20and%20heterogeneous%20databases%20used%20in%20bioinformatics.%22%2C%22date%22%3A%22May%202008%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.1093%5C%2Fbib%5C%2Fbbm064%22%2C%22ISSN%22%3A%221477-4054%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpubmed%5C%2F18192302%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-06T20%3A13%3A10Z%22%7D%7D%2C%7B%22key%22%3A%22W2P99XAZ%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Kocsor%20et%20al.%22%2C%22parsedDate%22%3A%222008%22%2C%22numChildren%22%3A3%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EKocsor%2C%20A.%2C%20Busa-Fekete%2C%20R.%2C%20%26amp%3B%20Pongor%2C%20S.%20%282008%29.%20Protein%20classification%20based%20on%20propagation%20of%20unrooted%20binary%20trees.%20%3Ci%3EProtein%20and%20Peptide%20Letters%3C%5C%2Fi%3E%2C%20%3Ci%3E15%3C%5C%2Fi%3E%285%29%2C%20428%26%23x2013%3B434.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.2174%5C%2F092986608784567492%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.2174%5C%2F092986608784567492%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22Protein%20classification%20based%20on%20propagation%20of%20unrooted%20binary%20trees%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Andr%5Cu00e1s%22%2C%22lastName%22%3A%22Kocsor%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22R%5Cu00f3bert%22%2C%22lastName%22%3A%22Busa-Fekete%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%5D%2C%22abstractNote%22%3A%22We%20present%20two%20efficient%20network%20propagation%20algorithms%20that%20operate%20on%20a%20binary%20tree%2C%20i.e.%2C%20a%20sparse-edged%20substitute%20of%20an%20entire%20similarity%20network.%20TreeProp-N%20is%20based%20on%20passing%20increments%20between%20nodes%20while%20TreeProp-E%20employs%20propagation%20to%20the%20edges%20of%20the%20tree.%20Both%20algorithms%20improve%20protein%20classification%20efficiency.%22%2C%22date%22%3A%222008%22%2C%22language%22%3A%22eng%22%2C%22DOI%22%3A%2210.2174%5C%2F092986608784567492%22%2C%22ISSN%22%3A%220929-8665%22%2C%22url%22%3A%22http%3A%5C%2F%5C%2Fwww.ncbi.nlm.nih.gov%5C%2Fpubmed%5C%2F18537730%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-06T20%3A06%3A56Z%22%7D%7D%2C%7B%22key%22%3A%22ZMJY9KY8%22%2C%22library%22%3A%7B%22id%22%3A2542344%7D%2C%22meta%22%3A%7B%22creatorSummary%22%3A%22Vera%20et%20al.%22%2C%22parsedDate%22%3A%222013-01-01%22%2C%22numChildren%22%3A2%7D%2C%22bib%22%3A%22%3Cdiv%20class%3D%5C%22csl-bib-body%5C%22%20style%3D%5C%22line-height%3A%202%3B%20padding-left%3A%201em%3B%20text-indent%3A-1em%3B%5C%22%3E%5Cn%20%20%3Cdiv%20class%3D%5C%22csl-entry%5C%22%3EVera%2C%20R.%2C%20Perez-Riverol%2C%20Y.%2C%20Perez%2C%20S.%2C%20Ligeti%2C%20B.%2C%20Kert%26%23xE9%3Bsz-Farkas%2C%20A.%2C%20%26amp%3B%20Pongor%2C%20S.%20%282013%29.%20JBioWH%3A%20an%20open-source%20Java%20framework%20for%20bioinformatics%20data%20integration.%20%3Ci%3EDatabase%3C%5C%2Fi%3E%2C%20%3Ci%3E2013%3C%5C%2Fi%3E%2C%20bat051.%20%3Ca%20href%3D%27https%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fdatabase%5C%2Fbat051%27%3Ehttps%3A%5C%2F%5C%2Fdoi.org%5C%2F10.1093%5C%2Fdatabase%5C%2Fbat051%3C%5C%2Fa%3E%3C%5C%2Fdiv%3E%5Cn%3C%5C%2Fdiv%3E%22%2C%22data%22%3A%7B%22itemType%22%3A%22journalArticle%22%2C%22title%22%3A%22JBioWH%3A%20an%20open-source%20Java%20framework%20for%20bioinformatics%20data%20integration%22%2C%22creators%22%3A%5B%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Roberto%22%2C%22lastName%22%3A%22Vera%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Yasset%22%2C%22lastName%22%3A%22Perez-Riverol%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Sonia%22%2C%22lastName%22%3A%22Perez%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Bal%5Cu00e1zs%22%2C%22lastName%22%3A%22Ligeti%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22Attila%22%2C%22lastName%22%3A%22Kert%5Cu00e9sz-Farkas%22%7D%2C%7B%22creatorType%22%3A%22author%22%2C%22firstName%22%3A%22S%5Cu00e1ndor%22%2C%22lastName%22%3A%22Pongor%22%7D%5D%2C%22abstractNote%22%3A%22Abstract.%20%20The%20Java%20BioWareHouse%20%28JBioWH%29%20project%20is%20an%20open-source%20platform-independent%20programming%20framework%20that%20allows%20a%20user%20to%20build%20his%5C%2Fher%20own%20integrate%22%2C%22date%22%3A%222013%5C%2F01%5C%2F01%22%2C%22language%22%3A%22en%22%2C%22DOI%22%3A%2210.1093%5C%2Fdatabase%5C%2Fbat051%22%2C%22ISSN%22%3A%22%22%2C%22url%22%3A%22https%3A%5C%2F%5C%2Facademic.oup.com%5C%2Fdatabase%5C%2Farticle%5C%2Fdoi%5C%2F10.1093%5C%2Fdatabase%5C%2Fbat051%5C%2F337984%22%2C%22collections%22%3A%5B%5D%2C%22dateModified%22%3A%222020-08-06T19%3A40%3A34Z%22%7D%7D%5D%7D Kertész-Farkas, A., Kocsor, A., & Pongor, S. (2009). The Application of Data Compression-Based Distances to Biological Sequences. In F. Emmert-Streib & M. Dehmer (Eds.),
Information Theory and Statistical Learning (pp. 83–100). Springer US.
https://doi.org/10.1007/978-0-387-84816-7_4 Kuznetsov, V., Lee, H. K., Maurer-Stroh, S., Molnár, M. J., Pongor, S., Eisenhaber, B., & Eisenhaber, F. (2013). How bioinformatics influences health informatics: usage of biomolecular sequences, expression profiles and automated microscopic image analyses for clinical needs and public health.
Health Information Science and Systems ,
1 (1), 2.
https://doi.org/10.1186/2047-2501-1-2 Busa-Fekete, R., Kertész-Farkas, A., Kocsor, A., & Pongor, S. (2008). Balanced ROC (BaROC) analysis for portien classification.
Journal of Biochemical and Biophysical Methods ,
70 (6), 1210–1214.
http://www.ncbi.nlm.nih.gov/pubmed/17689617 Kuzniar, A., van Ham, R. C. H. J., Pongor, S., & Leunissen, J. A. M. (2008). The quest for orthologs: finding the corresponding gene across genomes.
Trends in Genetics: TIG ,
24 (11), 539–551.
https://doi.org/10.1016/j.tig.2008.08.009 Kertész-Farkas, A., Dhir, S., Sonego, P., Pacurar, M., Netoteia, S., Nijveen, H., Kuzniar, A., Leunissen, J. A. M., Kocsor, A., & Pongor, S. (2008). Benchmarking protein classification algorithms via supervised cross-validation.
Journal of Biochemical and Biophysical Methods ,
70 (6), 1215–1223.
https://doi.org/10.1016/j.jbbm.2007.05.011 Sonego, P., Kocsor, A., & Pongor, S. (2008). ROC analysis: applications to the classification of biological sequences and 3D structures.
Briefings in Bioinformatics ,
9 (3), 198–209.
https://doi.org/10.1093/bib/bbm064 Kocsor, A., Busa-Fekete, R., & Pongor, S. (2008). Protein classification based on propagation of unrooted binary trees.
Protein and Peptide Letters ,
15 (5), 428–434.
https://doi.org/10.2174/092986608784567492 Vera, R., Perez-Riverol, Y., Perez, S., Ligeti, B., Kertész-Farkas, A., & Pongor, S. (2013). JBioWH: an open-source Java framework for bioinformatics data integration.
Database ,
2013 , bat051.
https://doi.org/10.1093/database/bat051